New Tools for Nanoscience Discovery
Understanding biological function and regulation requires characterising biomolecules, and their structures and interactions. Developing new ways to study biomolecules is a major focus of our research, and the tools we develop underpin much of our work. This research is enabled by the close collaboration between physical and life scientists that occurs at the Kavli.
Our extensive research in this area centres on developing new tools using mass spectrometry, next-generation imaging, novel bioanalytic technologies, and biosensors and probes. In addition to their scientific impacts, some of these tools have been successfully commercialised.
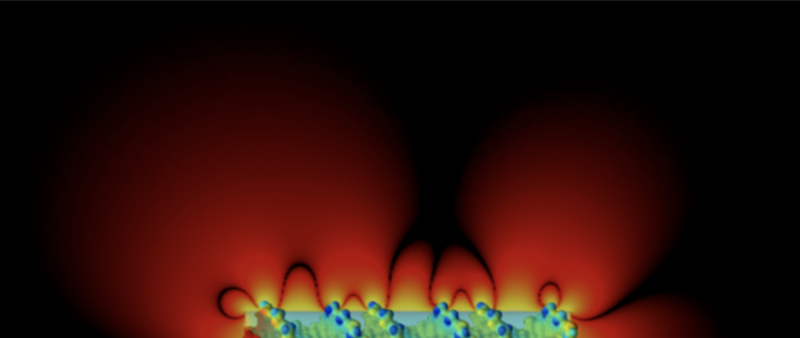
Difference in electrical potential surrounding the DNA double helix and an equivalent charged cylinder.
Mass spectrometry (MS) is one of the most powerful and widely used techniques for biophysical characterisation. The workhorse of proteomics, where it is relied on to identify proteins, it precisely measures the mass of peptides, proteins and other molecules. MS has applications in drug discovery and testing, biomarker screening, and many other areas.
Our researchers are renowned for pushing the boundaries of MS analysis – enabling more effective characterisation of protein assemblies and complexes as well as membrane proteins and their interactors, and identification of post-translational modifications; and for coupling MS with complementary technologies to gain deeper insights into protein structure and dynamics.
We also maintain world-class MS facilities.
Our work in this area focuses on:
-
Developing methods to eject membrane protein complexes intact from membranes for MS analysis – to study the complexes’ native behaviour and interactions (Carol Robinson)
-
Developing methods to combine ‘omics approaches with native MS, and identify possible regulators of membrane protein function (Carol Robinson)
-
Replicating membrane environments with novel detergents and membrane mimetics – to facilitate the study of membrane proteins (Carol Robinson)
-
Combining NMR spectroscopy or transmission EM with ion mobility MS to investigate structure as well as oligomeric states and associated dynamics (Andrew Baldwin, Justin Benesch)
-
Developing and applying MS-based techniques to characterise the structure, dynamics, and interactions of molecular chaperones and their targets (Justin Benesch)
-
Applying advanced MS methods and glycoprotein engineering to study glycoprotein dynamics and glycan-dependent interactions. (Weston Struwe)
-
Monitoring cancer progressing through changes in membrane protein transporters. (Carol Robinson)

Professor Robinson using a Mass Spectrometry instrument.
Imaging techniques, such as electron and fluorescence microscopy, enable the visualisation and study of phenomena at the nanoscale. Advances in imaging technology – including optics, probes, cameras and computational approaches – make it possible to investigate biomolecular structure, interactions and functional mechanisms at ever-increasing levels of detail.
Kavli researchers are developing and applying methods in a wide range of imaging modalities – including super-resolution fluorescence microscopy, single-molecule imaging, electron microscopy and digital holographic microscopy. Many of these methods focus on investigating single-molecule dynamics, such as the motion of molecular motors, the activity of chromatin during DNA replication, and the assembly and motion of a DNA translocase. Others enable the high-resolution 3D imaging of biomolecules within cells (cryo-ET) or imaging on lipid bilayers.
We are home to a cryo-EM facility. In addition, the neighbouring Micron Bioimaging Facility provides access to advanced imaging technology including the Nanoimager – a super-resolution fluorescence microscope developed at Oxford by Kavli INsD researchers for use in non-specialist laboratories.
Our work in this area focuses on developing:
-
Single-molecule fluorescence methods, instruments and assays – e.g. single-molecule FRET methods to study nanoscale motions while proteins function on their DNA substrates (Achillefs Kapanidis)
-
Single-molecule tracking and super-resolution imaging of proteins inside live single bacterial cells (Achillefs Kapanidis)
-
Electron cryotomography (cryoET) methods to directly image biomolecules within cells and cellular communities (Lindsay Baker)
-
Techniques in lipid bilayer systems for single-molecule microscopy (Richard Berry)
-
Single-molecule microscopy and biophysics techniques to study the mechanisms of the bacterial flagellar motor and other rotary molecular motors (Richard Berry)
-
Novel single-molecule imaging methods and applying them, plus other methods, to study mechanisms of DNA replication in eukaryotes (Dominika Gruszka)
-
Methods to handle and image large, fragile biomolecules with atomic precision (Stephan Rauschenbach)
-
Techniques in digital holographic microscopy (Richard Berry)
Kavli researchers are developing an array of innovative biophysical technologies, beyond mass spectrometry and imaging, for characterising biomolecules and biological systems. One example is mass photometry, which uses light scattering to measure the mass of individual proteins and other biomolecules. Another example is single-molecule electrometry, which provides a way to precisely measure the charge of biomolecules.
Our work in this area focuses on:
-
Exploring ultrafast mechano-chemistry occurring during the collision of large ions with surfaces at hyperthermal energies (Stephan Rauschenbach)
-
Expanding the capabilities and applications of mass photometry – a single-molecule mass measurement technology (Philipp Kukura)
-
Integrating single-molecule bioanalytical technologies for proteomics analysis (Justin Benesch, Philipp Kukura)
- Developing computational tools for the functional annotation of ion channel and nanopore structures (Stephen Tucker)
An image of a Mass Photometer.
Another important dimension of our work is the design and development of nanoscale biosensors and probes, nanostructures, and associated assays. These have applications in discovery science as well as the potential for use in clinical diagnostics and other fields.
Our work in this area focuses on:
-
Developing sensitive fluorescence imaging assays to detect pathogenic bacteria and the presence of antibiotic resistance with the help of machine learning (Achillefs Kapanidis)
-
Using selective nanobody probes to study K2P ion channels (Stephen Tucker)
Nuclear Magnetic Resonance Spectroscopy (NMR) allows the structure and dynamics of biomolecules to be characterised at atomic resolution. It allows interactions to be followed between molecules inside cells in solution in real-time, without the need for additional chemical labels such as fluorophores. The Kavli centre benefits from close proximity to high field NMR spectrometers, including an instrument operating at 22 Tesla, one of the highest field NMR spectrometers in the UK, and access to an instrument that can operate under biological safety level 2 conditions for studies of pathogens.
Our work in this area includes:
-
Developing new methods to study the strength and structures of interactions between small molecules (drugs) proteins both in isolation, and inside cells (Andrew Baldwin).
-
Development of methods to follow the mechanisms that allow proteins to aggregate to form amyloid in neurodegenerative diseases, and phase separate during cell development (Andrew Baldwin).
-
Development of new labelling schemes to take advantage of new ways to follow not only changes in structure, but also the dynamics of biomolecules inside cells (Andrew Baldwin, Justin Benesch, Weston Struwe, Stephan Rauschenbach).



